However, environmental conditions within Earth’s subsurface such as moisture, nutrient availability, and pH can significantly vary on incredibly small scales. Even at very small resolutions, these different environmental niches can largely impact the structure, function, and genomic potential of microorganisms – which can affect the important role microbes play in carbon and nutrient cycling.
A recent paper in ISME Communications led by Berkeley Lab Ecology Department Head Romy Charkaborty’s group aimed to study how these environmental niches shaped the genomic potential of bacteria in the genus Arthrobacter, which are commonly found in the soil and subsurface, and are known for their ability to break down larger and harder to degrade carbon molecules. To better understand and model the carbon cycle, scientists must study how changes in small-scale environmental conditions affect carbon metabolism and other cellular functions in closely related microbes (connect ecotypes to phenotypes and genotypes).
“It’s important to investigate how niche micro-environments influence microbes because microbes evolve and adapt to these environments, and will transform carbon differently,” explained lead author Sara Gushgari-Doyle, who performed this research while serving as an EESA postdoc fellow in Chakraborty’s lab and is now a scientist at ZymoChem. “Understanding these differences could even help us design strategies to predict and even manipulate the carbon cycle.”
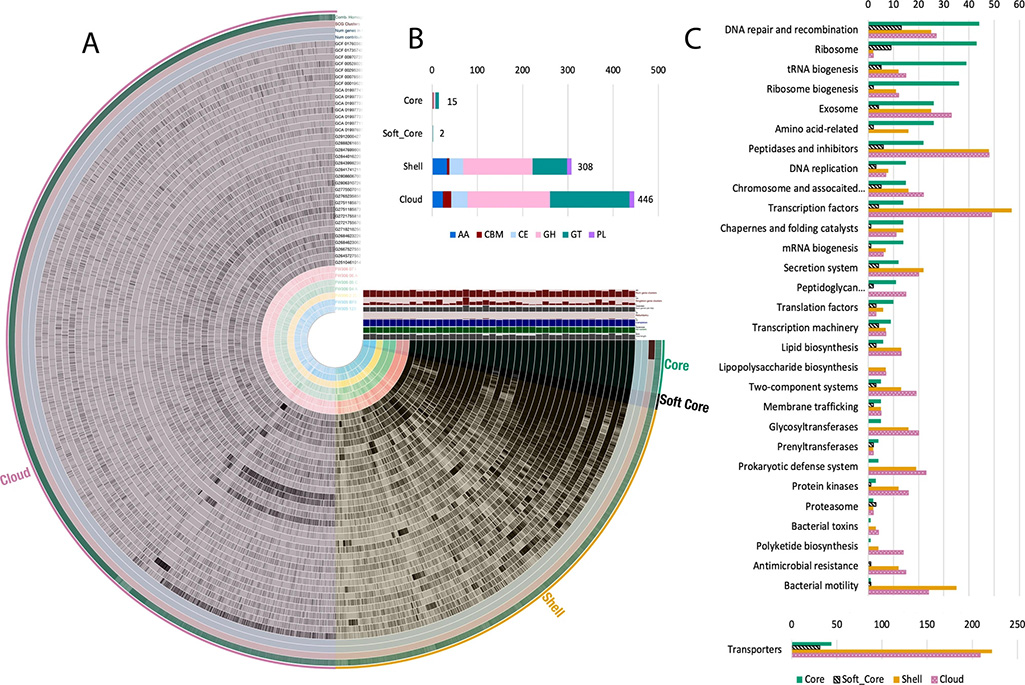
The team, which included members from Adam Arkin’s group in the Environmental Genomics and Systems Biology division at Berkeley Lab and Michael Adams’ group at the University of Georgia, isolated and cultivated seven distinct strains belonging to the genus Arthrobacter from varying depths of a single sediment core and associated groundwater from an adjacent well. These Arthrobacter isolates demonstrated functional and genomic capacities specific to the biogeochemical conditions of where they were sampled. They found that microbial strains, despite close spatial origin and phylogenetic relationships, break down carbon differently and had different sets of genes for carbon degradation. Further study of their genomes, along with 40 other Arthrobacter strains, revealed that the genus can indefinitely increase its set of genes and shows adaptability to available complex carbon substrates.
Understanding how very closely related bacteria transform carbon differently at small scales can advance global carbon cycle models and predictions of how microbes may affect the carbon cycle in a changing climate.